The full article can be downloaded here.
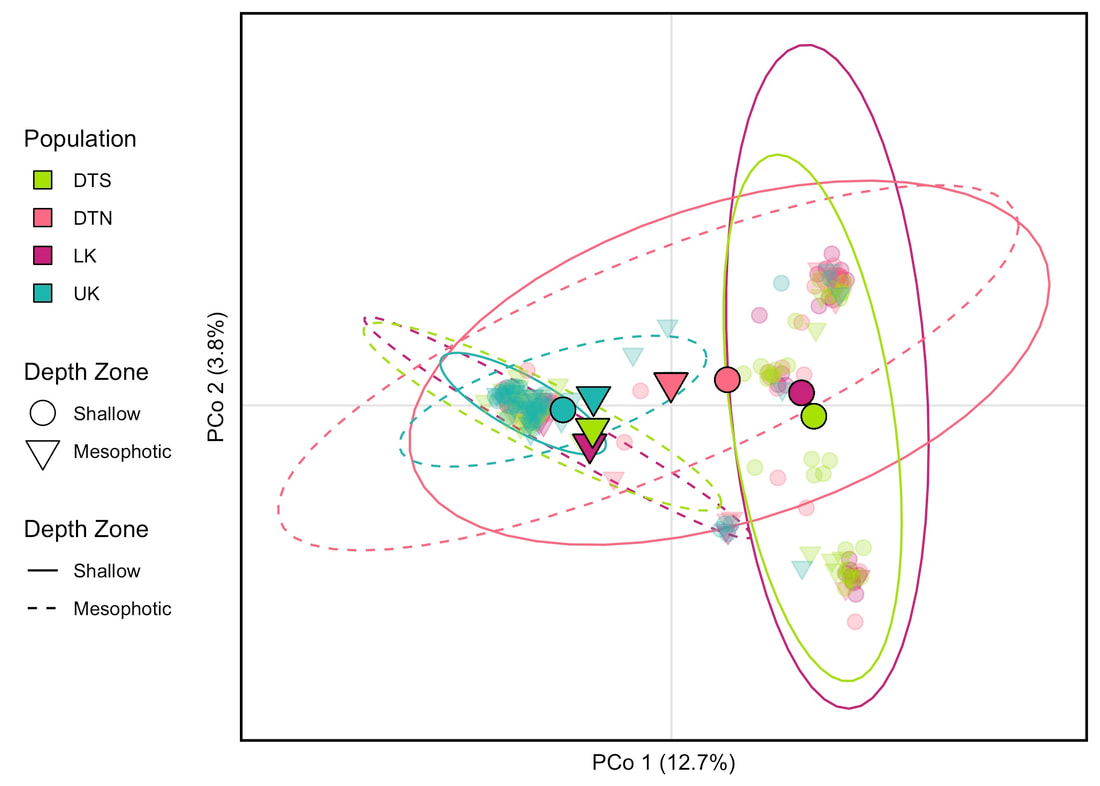
The study stemmed from a collaborative effort among Harbor Branch researchers to investigate the connectivity and health of shallow and mesophotic coral reefs across the Florida Keys National Marine Sanctuary. On a 2019 joint ROV and technical diving mission based off of the RV FG Walton Smith, the Voss lab led ROV, shallow, and technical diving teams collecting samples of multiple benthic invertebrate species. Species targets included the scleractinian coral Montastraea cavernosa from paired shallow (0-30 m) and upper mesophotic (30-150 m) depth zones in the Northern and Southern Dry Tortugas, Lower Keys, and Upper Keys. These samples were then genotyped using a 2bRAD SNP library preparation approach that generated over 9,000 SNP loci.
This work was funded by awards from NOAA Office of Ocean Exploration and Research to the Cooperative Institute for Ocean Exploration, Research and Technology (CIOERT) at FAU Harbor Branch and student funding from the NSF GRFP, Women Divers Hall of Fame, and Florida Sea Grant Scholars program. We are grateful to all of our Harbor Branch colleagues, Jake Emmert (Moody Gardens), the Undersea Vehicle Program at University of North Carolina at Wilmington, the crew of the RV FG Walton Smith, and the University of Texas at Austin’s Genome Sequencing and Analysis Facility who participated or facilitated this collaborative research expedition and follow-up molecular analyses. Special thanks go to the co-authors of the study including fellow Voss Lab Members: PhD student, Ryan Eckert and MS student, Ashley Carreiro.